
Using cross_validate in sklearn, simply explained
Cross_validate is a common function to use during the testing and validation phase of your machine learning model development. In this post I will explain what it is, what you can use it for, and how to implement it in Python.
Cross_validate is a common function to use during the testing and validation phase of your machine learning model development. In this post I will explain what it is, what you can use it for, and how to implement it in Python.
Cross_validate in sklearn, what is it?
Cross_validate is a function in the scikit-learn package which trains and tests a model over multiple folds of your dataset. This cross validation method gives you a better understanding of model performance over the whole dataset instead of just a single train/test split.
The process that cross_validate uses is typical for cross validation and follows these steps:
- The number of folds is defined, by default this is 5
- The dataset is split up according to these folds, where each fold has a unique set of testing data
- A model is trained and tested for each fold
- Each fold returns a metric/s for it's test data
- The mean and standard deviation of these metrics can then be calculated to provide a single metric for the process
An illustration of how this works is shown below:
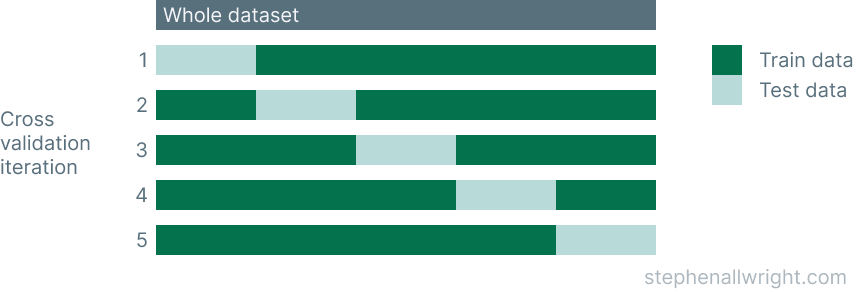
What is cross_validate used for?
Cross_validate is used as a cross validation technique to prevent over-fitting and promote model generalisation.
The typical process of model development is to train a model on one fold of data and then test on another. But how do we know that this single test dataset is representative? This is why we use cross_validate and cross validation more generally, to train and test our model on multiple folds such that we can be sure out model generalises well across the whole dataset and not just a single portion.
If we see that the metrics for all folds in cross_validate are uniform then it can be concluded that the model is able to generalise, however if there are significant differences between them then this may indicate over-fitting to certain folds and would need to be investigated further.
How many folds should I use in cross_validate?
By default cross_validate uses a 5-fold strategy, however this can be adjusted in the cv parameter.
But how many folds should you choose?
There is unfortunately no hard and fast rules when it comes to how many folds you should choose. A general rule of thumb though is that the number of folds should be as large as possible such that each fold has enough observations to generalise from and be tested on.
Can I train my model using cross_validate?
A common question developers have is whether cross_validate can also function as a way of training the final model. Unfortunately this is not the case. Cross_validate is a way of assessing a model and it’s parameters, and cannot be used for final training. Final training should take place on all available data and tested using a set of data that has been held back from the start.
Can I use cross_validate for classification and regression?
Cross_validate is a function which can be used for both classification and regression models. The only major difference between the two is that by default cross_validate uses Stratified KFold for classification, and normal KFold for regression.
Which metrics can I use in cross_validate?
By default cross_validate uses the chosen model’s default scoring metric, but this can be overridden in the scoring parameter. This parameter can accept either a single metric or multiple as a list or a dictionary.
The common metrics available in sklearn are passable as a string into this parameter, where some typical choices would be:
- ‘accuracy’
- ‘balanced_accuracy’
- ‘roc_auc’
- ‘f1’
- ‘neg_mean_absolute_error’
- ‘neg_root_mean_squared_error’
- ‘r2’
How to implement cross_validate in Python
This function is simple to implement in Python, but first let’s look at how it fits into a typical machine learning development workflow:
- Create a dataset
- Run hyper-parameter tuning
- Create model object with desired parameters
- Run cross_validate to test model performance
- Train final model on full dataset
Therefore, in order to use this function we need to first have an idea of the model we want to use and a prepared dataset to test it on. Let’s look at how this process would look in Python using a Linear Regression model and the Diabetes dataset from sklearn. We will also use a list of multiple metrics for the scoring parameter.
from sklearn import datasets
from sklearn.model_selection import cross_validate
from sklearn.linear_model import LinearRegression
X, y = datasets.load_diabetes(return_X_y=True)
metrics = ['neg_mean_absolute_error', 'r2']
model = LinearRegression()
scores = cross_validate(model, X, y, cv=5, scoring=metrics)
mae_scores = scores['test_neg_mean_absolute_error']
r2_scores = scores['test_r2']
print("Mean mae of %0.2f with a standard deviation of %0.2f" % (mae_scores.mean(), mae_scores.std()))
print("Mean r2 of %0.2f with a standard deviation of %0.2f" % (r2_scores.mean(), r2_scores.std()))
Function parameters for cross_validate
There are a number of parameters that you should be aware of when using this function. They are:
estimator- The model object to use to fit the dataX- The data to fit the model ony- The target of the modelscoring- The error metric/s to usecv- The number of splits to use
Summary of the cross_validate function
Cross_validate is a method which runs cross validation on a dataset to test whether the model can generalise over the whole dataset. The function returns a list of scores per fold, and the average of these scores can be calculated to provide a single metric value for the dataset. This is a function and a technique which you should add to your workflow to make sure you are developing highly performant models.
Related articles
Using cross_val_score in sklearn
Difference between cross_val_score and cross_validate
Fix sklearn.cross_validation import error
What is a baseline machine learning model?
References
cross_validate documentation
Cross validation guide from sklearn
